Is rheumatoid arthritis genetic
If you tested your DNA with a personal genomics service like 23andMe, AncestryDNA, FamilyTreeDNA, MyHeritage or another testing company, you can learn more about your risk factors for hundreds of diseases. By clicking the button above ⬆️, you can upload your raw DNA data file and receive a personalized 250-page health report with research links that is the most comprehensive.
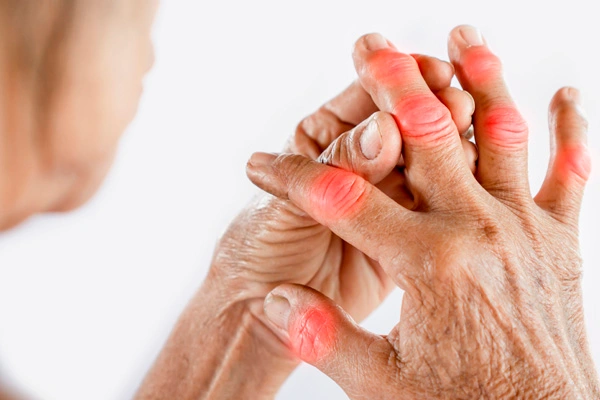
Chronic abnormal inflammation is the hallmark of rheumatoid arthritis, a disease that primarily affects the joints. The most common symptoms include joint pain, swelling, and stiffness, with small joints in the hands and feet being the most frequently affected. However, larger joints like the shoulders, hips, and knees may also become involved later in the disease. The pattern of joint involvement is typically symmetrical, meaning that if one hand is affected, the other hand is likely to be involved as well. People with rheumatoid arthritis often experience increased joint pain and stiffness upon waking up in the morning or after prolonged periods of rest.
Inflammation of various tissues and organs, such as the eyes, lungs, and blood vessels, can also be caused by rheumatoid arthritis. Other indications and manifestations of this ailment may comprise fatigue, mild fever, weight loss, and anemia due to a deficiency of red blood cells. Certain individuals may also develop rheumatoid nodules, which are solid, benign growths that can emerge beneath the skin or in other parts of the body.
Numerous genes have been analyzed to determine their potential role as risk factors for rheumatoid arthritis, with a majority of them being associated with immune system function. The most noteworthy genetic risk factors for this condition are variations in the human leukocyte antigen (HLA) genes, particularly the HLA-DRB1 gene. These genes produce proteins that aid the immune system in distinguishing between the body's own proteins and those produced by foreign invaders like viruses and bacteria. Other gene variations seem to have a lesser effect on an individual's likelihood of developing rheumatoid arthritis.
STAT4, TRAF1/C5, and PTPN22 are genetic markers associated with rheumatoid arthritis. The STAT4 gene is responsible for regulating and activating the immune system, and mutations in this gene are also found in autoimmune disorders such as lupus. The TRAF1/C5 genes are known to contribute significantly to chronic inflammation. Additionally, in Caucasian patients, the PTPN22 gene plays a crucial role in supporting immune cell responses, affecting the development and manifestation of RA. This gene is one of the top genes linked to the risk of developing RA.
HLA-DR4, also referred to as Human Leukocyte Antigen or the major histocompatibility complex (MHC), is a gene that distinguishes itself from others. According to Xinli Hu, MD, PhD, director of computational genetics and a senior computational geneticist in Systems Immunology at Pfizer, this gene is predominantly linked with RA. Individuals possessing this gene have a higher likelihood of developing RA compared to those who do not.
Rheumatoid arthritis is thought to be influenced by non-genetic factors as well. Although the exact mechanism is unclear, these factors may activate the condition in individuals who are susceptible. Some potential triggers include alterations in sex hormones, particularly in women, exposure to specific types of dust or fibers in the workplace, and viral or bacterial infections. Additionally, long-term smoking is a known risk factor for developing rheumatoid arthritis and is linked to more severe symptoms in those who already have the disease.
Follow the link of the selected polymorphism to read a brief description of how the selected polymorphism affects Rheumatoid arthritis and see a list of existing studies.
SNP polymorphisms related to the topic Rheumatoid arthritis:
| rs2476601 | This important SNP, located in the PTPN22 gene and also known as R620W or 1858C>T, may influence the risk of multiple autoimmune diseases such as rheumatoid arthritis, type 1 diabetes, autoimmune thyroiditis and systemic lupus erythematosus. |
| rs5029937 | The single nucleotide polymorphism rs5029937 in the TNFAIP3 gene correlates with the risk of rheumatoid arthritis. |
| rs10818488 | The rs10818488 polymorphism in the TRAF1/C region is associated with genetic predisposition to rheumatoid arthritis and systemic lupus erythematosus. |
| rs3738919 | The ITGAV rs3738919-C allele is associated with rheumatoid arthritis in Caucasians. |
| rs10488631 | The interferon regulatory factor 5 (IRF5) gene variant causes a 2-fold increased risk of systemic lupus erythematosus |
| rs2736340 | The FAM167A-BLK rs2736340 polymorphism is associated with susceptibility to autoimmune diseases, particularly rheumatoid arthritis and systemic lupus erythematosus. |
| rs3087243 | The CTLA4 allelic variant alters T cell phosphorylation patterns and causes an increased risk of autoimmune diseases. |
| rs11676922 | The combination of CD28 (rs1980422) and IRF5 (rs10488631) polymorphisms is associated with seropositivity in rheumatoid arthritis. |
| rs26232 | The C5orf30 rs26232 variant is a negative regulator of tissue damage in rheumatoid arthritis and is associated with joint damage in rheumatoid arthritis. |
| rs874040 | RBPJ polymorphism associated with rheumatoid arthritis alters memory CD4+ T cells. |
| rs2230926 | Multiple polymorphisms in the TNFAIP3 region are independently associated with rheumatoid arthritis and systemic lupus erythematosus. |
| rs2104286 | Genetic heterogeneity of IL2RA indicates susceptibility to multiple sclerosis and susceptibility to type 1 diabetes. |
| rs13119723 | Combined with the rs6822844 breakage, the study showed the strongest association with celiac disease among Caucasian patients. |
| rs6822844 | Combined with the rs13119723 breakage, the study showed the strongest association with celiac disease among Caucasian patients. |
| rs6679677 | An allelic variant of the PHTF1 gene is associated with a 5.2-fold increased risk of type 1 diabetes and a 3.3-fold increased risk of rheumatoid arthritis. |
| rs3184504 | A variant of celiac disease genetic risk associated with immune response. Also carrier associated type 1 diabetes. |
| rs7574865 | 1.3-fold risk of rheumatoid arthritis |
| rs3218251 | |
| rs13315591 | |
| rs9372120 | |
| rs67250450 | |
| rs11761231 | |
| rs12529514 | |
| rs4409785 | |
| rs13142500 | |
| rs998731 | |
| rs10821944 | |
| rs12831974 | |
| rs13330176 | |
| rs11089637 | |
| rs4780401 | |
| rs231735 | |
| rs13031237 | |
| rs3093023 | |
| rs2867461 | |
| rs2327832 | |
| rs4750316 | |
| rs2561477 | |
| rs9826828 | |
| rs6920220 | |
| rs6732565 | |
| rs678347 | |
| rs657075 | |
| rs1877030 | |
| rs73013527 | |
| rs3824660 | |
| rs2736337 | |
| rs2469434 | |
| rs615672 | |
| rs6859219 | |
| rs10499194 | |
| rs71508903 | |
| rs6496667 | |
| rs2961663 | |
| rs6715284 | |
| rs726288 | |
| rs6457620 | |
| rs4452313 | |
| rs8133843 | |
| rs11574914 | |
| rs2664035 | |
| rs2847297 | |
| rs2240335 | |
| rs805297 | |
| rs947474 | |
| rs10865035 | |
| rs10774624 | |
| rs3761847 | |
| rs2671692 | |
| rs10985070 | |
| rs6457617 | |
| rs3816587 | |
| rs7528684 | |
| rs2837960 | |
| rs11889341 | |
| rs3890745 | |
| rs2240340 | |
| rs1953126 | |
| rs743777 | |
| rs6684865 | |
| rs9550642 | |
| rs11162922 | |
| rs11203366 | |
| rs8032939 | |
| rs13192471 | |
| rs331463 | |
| rs2317230 | |
| rs1950897 | |
| rs1571878 | |
| rs3781913 | |
| rs4810485 | |
| rs4305317 | |
| rs3093024 | |
| rs12131057 | |
| rs9275406 | |
| rs8026898 | |
| rs1043099 | |
| rs2075876 | |
| rs2841277 | |
| rs1980422 | |
| rs34695944 | |
| rs2451258 | |
| rs3125734 | |
| rs1516971 | |
| rs11933540 | |
| rs227163 | |
| rs968567 | |
| rs72634030 | |
| rs11900673 | |
| rs881375 | |
| rs624988 | |
| rs2233424 | |
| rs9653442 | |
| rs28411352 | |
| rs73081554 | |
| rs2582532 | |
| rs1858037 | |
| rs909685 | |
| rs6910071 | |
| rs9571178 | |
| rs2812378 | |
| rs11203203 | |
| rs934734 | |
| rs660895 | |
| rs4272 | |
| rs13017599 | |
| rs3806624 | |
| rs10175798 | |
| rs1893592 | |
| rs45475795 | |
| rs951005 | |
| rs7765379 | |
| rs12140275 | |
| rs9268839 | |
| rs2233434 | |
| rs4678 | |
| rs12525220 | |
| rs7731626 | |
| rs13192841 | |
| rs2072438 | |
| rs1854853 | |
| rs12379034 | |
| rs3763309 | |
| rs1160542 | |
| rs1678542 | |
| rs10760130 | |
| rs3766379 | |
| rs2872507 | |
| rs6682654 | |
| rs2442728 | |
About The Author
Li DaliLi Dali, a National Foundation for Outstanding Youth Fund recipient, is a researcher at the School of Life Sciences in East China Normal University. He earned his PhD in genetics from Hunan Normal University in 2007 and conducted collaborative research at Texas A&M University during his doctoral studies. Li Dali and his team have optimized and innovated gene editing technology, leading to the establishment of a world-class system for constructing gene editing disease models.